Antibiotic-resistant bacteria already existed in the environment before antibiotics were used in human medicine.1 Besides promoting the development of new resistance traits, the way and extent we use antibiotics today in food production selects for existing resistant clones within microbial populations and promotes their expansion. The spread of antibiotic resistance genes and the development of multi-drug resistance represent major threats to public health.2,3
Varied antibiotics use
As antibiotics are not only used in human and veterinary medicine, but also in animal food production and agriculture,4,5 the environment plays an important role in the spread of antibiotic resistances, because it provides a multitude of relevant niches (soil, water, plants, animals) for the spread of resistance.
The evolution of antibiotic resistance and the interplay of clinical and environmental bacteria is not yet well understood. In 2001, the European Council recommendation on the prudent use of antimicrobial agents in human medicine supported the idea that the occurrence of antibiotic resistance in human pathogens correlates with their occurrence in animals and the environment.6 Against this background, the European Food Safety Authority (EFSA) has re-evaluated antibacterial products used as feed additives and their impact on resistance development to antibiotics of human and veterinary importance.7
In 2010, 63,200 tons of antibiotics were used in livestock and this number is expected to increase to 105,600 tons in the year 2030.8 It is easy to understand this excessive use of antibiotics promotes the emergence of antibiotic resistant pathogenic as well as commensal bacteria.
This process is a major concern not only for animal, but also for human health, as some of these bacteria have zoonotic potential or serve at least as a reservoir for resistance genes, which can be transferred to other bacteria including human pathogens. Based on a recent decision of the European Commission regarding the monitoring of antimicrobial resistance in zoonotic and commensal agents in food-producing animals and meat, the detection of changes in antibiotic resistance patterns in animal populations should help define future trends in the occurrence of antimicrobial resistance.9
The ongoing opposition
So far, attempts to prevent or counteract the occurrence and spread of antibiotic resistance in food animals were mainly based on a reduction of antibiotic use in food production. As a consequence, the use of antibiotics as growth promoters has been banned in the European Union.10
However, this decision has several unexpected consequences, such as an overall negative impact on animal health, animal morbidity and increased levels of certain pathogens in livestock, which was considered to be associated with increased numbers of food-borne infections in the EU.11
The biggest problem associated with the ban of antibiotics as growth promoters is the observed concomitant increase of therapeutic use of antimicrobial substances in animal production. Compared with the situation before the ban, the total amount of antibiotics used in conventional livestock breeding increased in some countries.11 As the demand for animal products will constantly rise due to the growing human population, this calls for further improved knowledge regarding the emergence and spread of antibiotic resistance traits in order to develop strategies to counteract antibiotic resistance.
Microbial antibiotic resistance
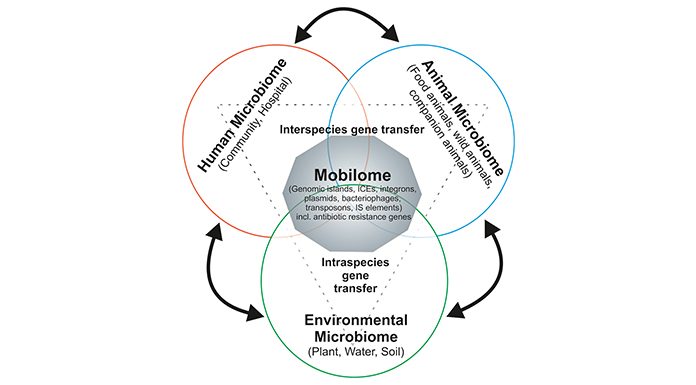
Microbial antibiotic resistance traits and individual mobile genetic elements (MGEs), on which resistance determinants are frequently located, have been intensively studied regarding the spread of antimicrobial drug resistance. However, the composition and dynamics of the global bacterial mobile gene pool in a given niche (humans, animals) or the environment, the so-called ‘mobilome’, has not been comprehensively studied so far. The mobilome consists of MGEs, including plasmids, bacteriophages, genomic islands (GEIs), integrative and conjugative elements (ICEs), integrons, and transposons. These MGEs can frequently carry antibiotic resistance genes and serve as vectors for the lateral dissemination of antibiotic resistance determinants between microbes.
Among MGEs, plasmids, which can carry (multiple) resistance determinants, are among the most important vehicles for the spread of antibiotic resistance. In order to better understand the constraints and driving forces of horizontal gene transfer (HGT), we have to determine the molecular basis of the bacterial host range of resistance plasmids, the contribution of environmental conditions to the transfer efficiency of such plasmids and the impact that resistance plasmid carriage has on the fitness and competitiveness of the recipient hosts.
The uptake and exchange of bacteria between the environment, animals and humans increases the diversity and dynamics of the mobilome. In the wake of metagenomic analyses, the prevalence of antibiotic resistance genes in the commensal microbiota as well as compositional changes of the microbiota are more and more in the research focus. Recent analyses indicate that resistance genes are widely distributed in the intestinal gut microbiota of healthy individuals.12-14 Accordingly, the detailed analysis of the intestinal mobilome, not only in humans, but also in companion and food animals is critical for our understanding of the spread of antibiotic resistance as well as for relevant preventive approaches.14-17
Although we are slowly beginning to elucidate (i) the impact of antibiotics on the composition of the microbiota and environmental microbial communities and (ii) the relevance of certain compositional changes of the microbiota regarding its impact on human health, our knowledge of the resistance mobilome in general, the interplay between different mobilome components and different members of the intestinal microbiota or environmental microbial communities is still limited.15,16,18-20
Accordingly, in order to improve tracking of antibiotic resistance, food safety and preventive as well as therapeutic health measures, we have to strengthen more holistic approaches including studies of the diversity, dynamics and evolution of antibiotic resistance genes and their vectors together with compositional analyses of their reservoirs in the environment, food animals and the human population.
References
-
- D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. (2011) Antibiotic resistance is ancient. Nature 477:457–461.
- McKenna M (2013) Antibiotic resistance: the last resort. Nature 499:394–396, 10.1038/499394a.
- World Health Organization (2015) Global action plan on antimicrobial resistance. Geneva, Switzerland.
- Marshall BM, Levy SB (2011) Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 24:718-733.
- Garcia-Alvarez L, Dawson S, Cookson B, Hawkey P. (2012) Working across the veterinary and human health sectors. J Antimicrob 67:i37-49.
- European Council (2001) Council Recommendation of 15.11.2001 on the Prudent Use of Antimicrobial Agents in Human Medicine (2002/77/EC). OJ L34 of 5.2.2002, p.13 European Council (EC), Brussels, Belgium.
- The European Food Safety Authority (EFSA) Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2012) Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J, 10:2740.
- United Nations, Department of Economic and Social Affairs, Population Division (2015) World Population Prospects: The 2015 Revision, Key Findings and Advance Tables. Working Paper No. ESA/P/WP.241.
- European Commission (2013) Commission Implementing Decision 613/2013 of 12.11.2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and comensal bacteria.
- European Commission (2003) Regulation 1831/2003/EC on additives for use in animal nutrition.
- Hao H, Cheng G, Iqbal Z, Ai X, Hussain HI, Huang L, Dai M, Wang Y, Liu Z, Yuan Z (2014) Benefits and risks of antimicrobial use in food-producing animals. Front Microbiol 5:288.
- Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, Pan Y, Li J, Zhu L, Wang X, Meng Z, Zhao F, Liu D, Ma J, Qin N, Xiang C, Xiao Y, Li L, Yang H, Wang J, Yang R, Gao GF, Wang J, Zhu B (2013) Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 4:2151.
- Sommer MOA, Dantas G, Church GM (2009) Functional Characterization of the Antibiotic Resistance Reservoir in the Human Microflora. Science 325:1128–1131.
- Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ (2011) Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480:241-244.
- Modi SR, Lee HH, Spina CS, Collins JJ (2013) Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499:219–222.
- Devirgiliis C, Barile S, Perozzi G (2011) Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 6:275–284.
- Sekirov I, Russell SL, Antunes LC, Finlay BB (2010) Gut microbiota in health and disease. Physiol Rev. 90:859-904.
- Cameron A, McAllister TA (2016) Antimicrobial usage and resistance in beef production. J Anim Sci Biotechnol. 7:68.
- Thiemann S, Smit N, Strowig T (2016) Antibiotics and the Intestinal Microbiome: Individual Responses, Resilience of the Ecosystem, and the Susceptibility to Infections. Curr Top Microbiol Immunol. 398:123-146.
- Marti E, Huerta B, Rodríguez-Mozaz S, Barceló D, Balcázar JL, Marcé R. (2016) Effects of subinhibitory ciprofloxacin concentrations on the abundance of qnrS and composition of bacterial communities from water supply reservoirs. Chemosphere. 161:470-4.